Tutorial
1. What is PAMDB?
PAMDB aims to bridge the gap between published circuits and lack of universal quantitive parameters for their parts. PAMDB includes parameter values inferred from hundreds of papers under a single unified model. PAMDB can be used to rank parts based on their activity, acquire parameter values and quantify their uncertainty, simulate circuits and visualize the part universe for the published synthetic biology work so far.
2. Why do we need PAMDB?
Mathematical modeling and numerical simulation are crucial to support design decisions in synthetic biology. For the models to be predictive rather than descriptive, it is important that the key processes are captured in the model and the modeling parameters are accurate enough so that the simulation outcome reflects reality. Currently, there is a plethora of models, most of them built to support experimental work, which are fitted to describe the observed data. Similarly, although some parameters are common, many are not, which makes interoperability difficult or impossible. Given the cost in time and expenses to experimentally measure them, parameter inference and easy access to a parameter repository for parts is important to move the field forward.
3. What information PAMDB holds and what is its structure?
3.1 Overview
Here, we have curated and integrated data from 45 publications that contain 118 circuits and 165 genetic parts of the bacterium Escherichia coli. We used a succinct, universal model formulation to describe the part behavior in each circuit. We introduce a constrained consensus inference method that was used to infer the value of the model parameters. This work provides a resource and a methodology that can be used as a point of reference for synthetic circuit modeling.
3.2 PAMDB structure
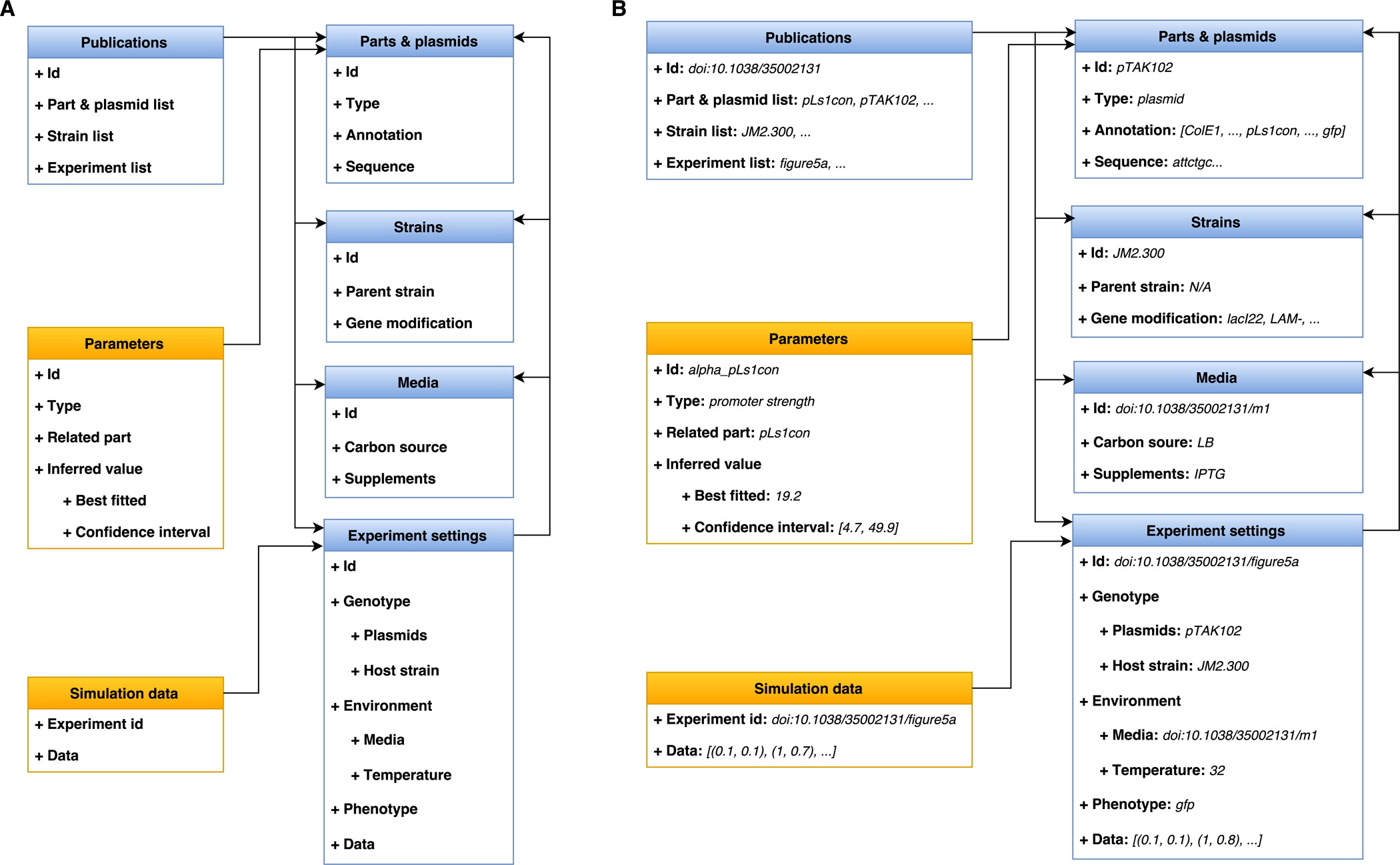
3.3 Model & parameter
To avoid adding ill-constrained parameters, we used a simple model based on Hill and linear functions. This model captured both the processes of transcription, translation and ligand-protein binding. More specifically, when a transcription factor TF is bound to a promoter Pr, the expression level νg of a gene g at the downstream of Pr is modeled by
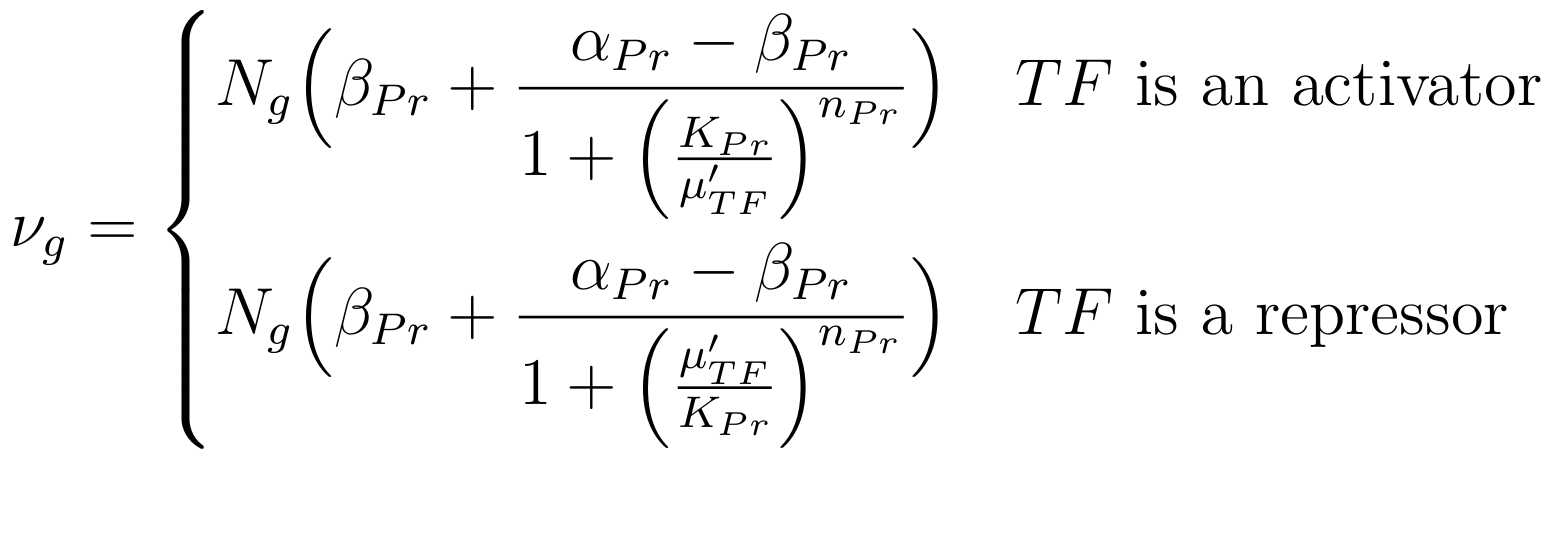
where Ng is the copy number of gene g (Ng is the plasmid copy number if g is on a plasmid, otherwise, Ng = 1 if g is in the chromosome). Parameters βPr, αPr, KPr, and nPr represent the basal level, the promoter strength, the binding affinity and its cooperativity, respectively, with respect to promoter Pr and its transcription factor TF. Here, μTF' is the concentration of transcription factor TF and is modeled as

where μTF is the protein expression level (from the equation below). [LTF] is the ligand concentration when a ligand LTF binds to the transcription factor LTF. Parameters KLTF and nLTF correspond to the dissociation constant and the Hill coefficient of the ligand, respectively. For the translation, the protein expression level μg of a gene g is modeled by
where αr is the strength of the ribosome binding site r in the upstream of g. In the case where g is a reporter protein (e.g., gfp, yfp), the protein expression level μg in relative units is converted to the protein expression level μgau in arbitrary units to fit with the experimental data. This is achieved by applying the following conversion formula
where ωgref is the scale factor for the reporter protein g as presented in publication ref. The value of this scale factor will be estimated simultaneously with the value of other parameters to fit the experimental data with the model through the parameter inference process
4. How to use PAMDB?
4.1 Find information of an item
- Search by it's name (currently, only part name can be searched)
- Find an item by it's category including publication , part , strain , experiment , or parameter
4.2 Download the dataset
Currently, the summary information of each category can be downloaded as a table (csv, excel or pdf). We are testing APIs to download the detail data of each item from the database.
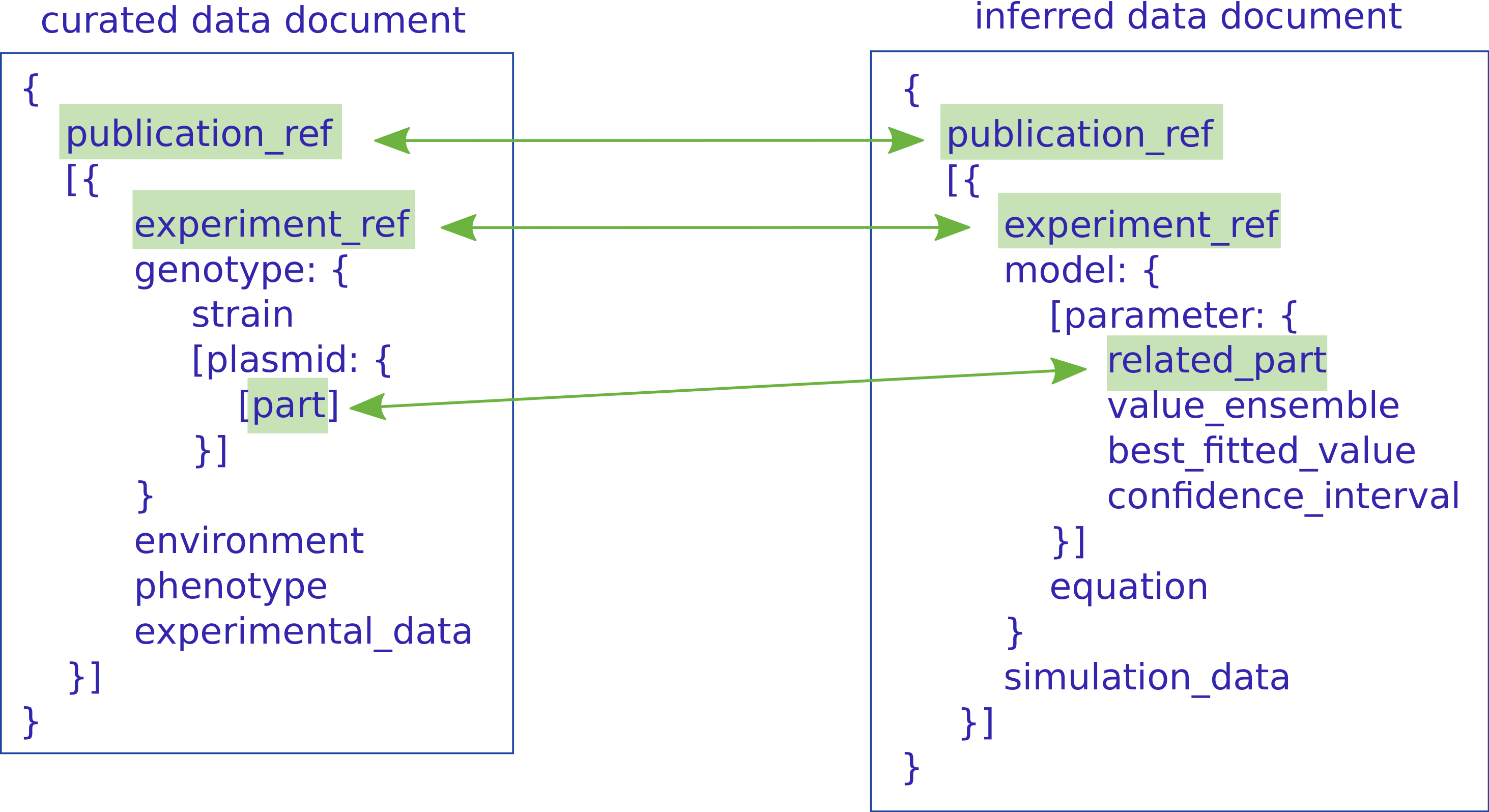
5. Update, maintenance, and comments
The current version is 1.0, we will list all changes from the previous version when the site is updated. Please contact us for any question, comment or suggestion.